Monitoring salmonella in the environment is critical because contaminated freshwater used to irrigate crops can transfer pathogenic bacteria to fresh produce, causing illness and even death. But University of Georgia researcher Nikki Shariat believes traditional salmonella surveillance methods have been missing the big picture.
Shariat, an assistant professor of population health at UGA’s College of Veterinary Medicine, has found that salmonella populations in freshwater are more diverse than previously thought. And that this diversity disguises hidden salmonella populations that are clinically important.
In a recent study published in Applied and Environmental Microbiology, Shariat, principal investigator on the project, detailed not only the change in salmonella populations in the Susquehanna River Basin over time but also uncovered hidden populations that have, until now, gone undetected.
Using a tool developed in her lab, CRISPR-SeroSeq, Shariat and her team of students used next-generation sequencing to dive deep and reveal entire populations of salmonella in a sample. In 78% of samples from the Susquehanna River Basin containing salmonella, the team discovered eight of the CDC’s top 10 human-illness-causing serotypes. Of note, six of these serotypes were often completely hidden by more abundant, less clinically relevant types of salmonella. In a traditional survey, these serotypes would have been missed entirely.
The Susquehanna River provides 70% of the freshwater to the Chesapeake Bay, and large swaths of it and its tributaries course through agricultural areas. During spring, the area receives increased amounts of rain that produce runoff that ultimately feeds the rivers and creeks that fill the Susquehanna.
It is this runoff that the researchers believe promotes the increased amount of salmonella found in samples collected in spring. Single samples contained up to 10 serotypes of salmonella—many of which are associated with animal agriculture.
“In food animals, we know that when salmonella grows, many serotypes survive. And in my work with poultry and other food animals here at UGA, we have seen multiple serotypes present in a single sample,” said Shariat. “But salmonella is adapted to grow in the intestines of animals, not freshwater. We wanted to see what diversity could be found in water.”
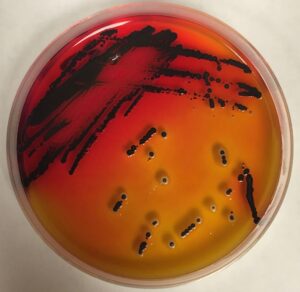
Typically, when an environment is surveyed for salmonella, samples are collected, cultured to enrich for salmonella, and then plated onto selective indicator agar plates. “On these plates, salmonella typically grows as distinctive black colonies,” Shariat said, “and individual colonies are sampled from the plate and characterized by their serotype.”
A researcher selects one or two colonies from the plate to characterize, which means that only the most abundant serotypes get identified. But, as Shariat’s research has continually found, this method only scratches the surface of a broad and diverse population of salmonella.
“Using the plate method, you might have to pick hundreds of separate colonies to find a trace of these background serotypes,” Shariat said. “This would be logistically impossible and very expensive.” Using CRISPR-SeroSeq, the team can classify and characterize all identifiable serotypes in a single sample. “What was impressive about our Susquehanna work is that over 80% of samples that contained salmonella actually included more than one type of salmonella.”
Student Abigail Deaven (left) and Nikki Shariat collect samples in the Susquehanna River. (Submitted photo taken in 2019)
In future studies, the team hopes to further investigate how long-lived salmonella is in freshwater and how far it can travel within a watershed.
This study was funded by the Pennsylvania Sea Grant and the National Oceanic and Atmospheric Administration and was performed in partnership with Juniata College and the U.S. Food and Drug Administration Center for Food Safety and Applied Nutrition. The Susquehanna River and its tributaries represent the largest subject for a study of salmonella presence and diversity.